目录
Motivation
Programming Language Independence Interface(PISA) provides many useful tools for multi-objective evolutionary algorithms. The ones that I am interested in are on quality assessment.
indicators
input format:
obj_val_1_run_1 obj_val_2_run_1 ...
obj_val_1_run_1 obj_val_2_run_1 ...
obj_val_1_run_1 obj_val_2_run_1 ...
[blank line]
obj_val_1_run_2 obj_val_2_run_2 ...
obj_val_1_run_2 obj_val_2_run_2 ...
obj_val_1_run_2 obj_val_2_run_2 ...output format:
indicator_run_1
indicator_run_2dominance-rank
./dominance-rank [<param_file>] <data_file1> <data_file2> <output_file>
<param_file> specifies the name of the parameter file for dominance-rank; the file has the following format:
dim <integer>
obj <+|-> <+|-> ...eps_ind
./eps_ind [<param_file>] <data_file> <reference_set> <output_file>
<param_file> specifies the name of the parameter file for eps_ind; the file has the following format:
dim <integer>
obj <+|-> <+|-> ...
method <0|1>additive(0), multiplicative(1)
hyp_ind
./hyp_ind [<param_file>] <data_file> <reference_set> <output_file>
<param_file> specifies the name of the parameter file for hyp_ind; the file has the following format:
dim <integer>
obj <+|-> <+|-> ...
method <0|1>
nadir <real> <real> ...relative to the reference set (1) or not (0)
r_ind
./r_ind [<paramfile>] <datafile> <referencefile> <outfile>
<paramfile> specifies the name of a parameter file which must have the following format:
dim <positive integer>
obj <-|+> <-|+> ...
ideal <double> <double> ...
nadir <double> <double> ...
rho <small positive double>
method <1|2|3>
R <{1|2|3>
s <positive integer>the weighted sum (1), Thebycheff (2), or augmented Thebycheff function (3)
dim=2, s=500; dim=3, s=30; dim=4, s=12; dim=5, s=8
statistics
fisher-indep
./fisher-indep <indicator_file> <param_file> <output_file>
<param_file> is the name of a file with precisely two lines, as follows:
nresamples 5000
seed 6756519834The output is the p-value of the one-tailed test for rejecting the null hypothesis of no difference between each pair of sample populations
fisher-matched
./fisher_matched <indicator_file> <paramfile> <outputfile>
<param_file> is the name of a file with precisely two lines, as follows:
nresamples 5000
seed 6756519834The output is the one-tailed p-value that the null hypothesis is true for each pair
kruskal-wallis
./kruskal <indicator_file> <param_file> <output_file>
<param_file> is the name of a file with the following one-line format
alpha 0.05If and only if a first test for significance of any differences between the samples is passed, at the given alpha value, then the output will be the one-taileed p-values for rejecting a null hypothesis of no significant difference between two samples, for each pair-wise combination. In the case that the first test fails, the output is simply, “H0”.
mann-whit
./mann-whit <indicator_file> <param_file> <output_file>
<param_file> is the name of a file that, at present, may be emptyThe output is the p-value of the two-tailed test for rejecting the null hypothesis of no difference between each pair of sample populations
wilcoxon-sign
./wilcoxon <indicator_file> <param_file> <output_file>
<param_file> is the name of a file that, at present, may be emptyThe output is the one-tailed p-value that the null hypothesis is true for each pair
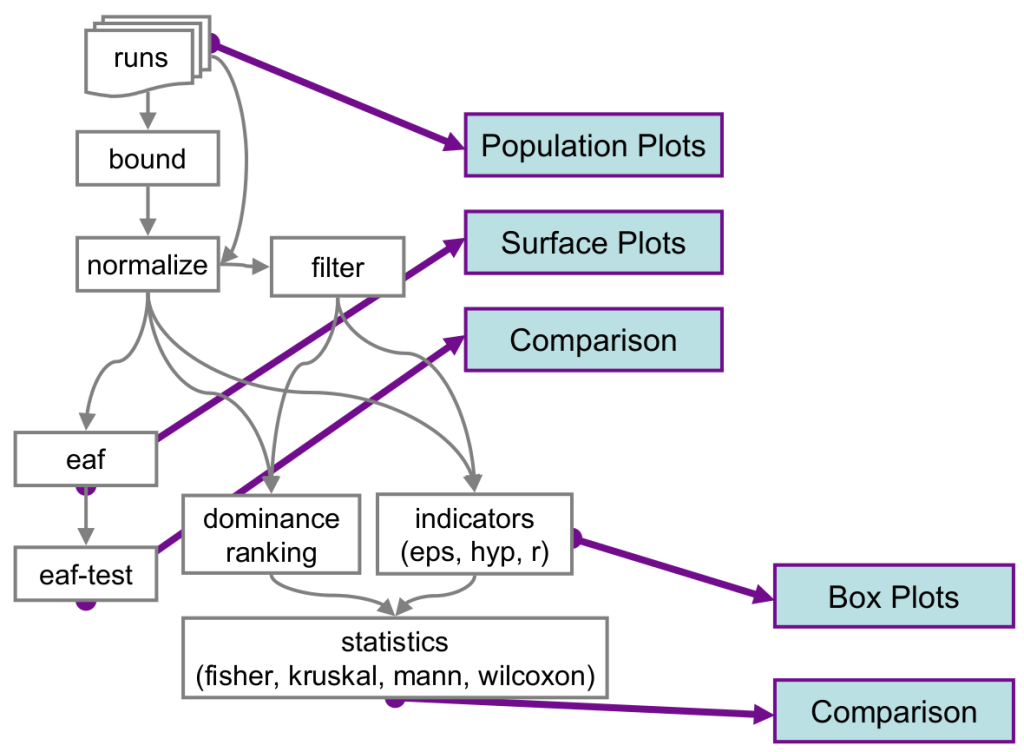
tools
bound
A program that reads in a file consisting of a collection of approximation sets, assumed to be from all the optimizers under comparison, and not necessarily internally nondominated sets. A separate parameter file gives the dimension of the data and whether each objective should be minimized or maximized. It also gives a parameter phi. The output is the upper and lower bound in each objective, the ideal point and the nadir point, and the latter when shifted by phi times the range (in each objective).
./bound [<param>] <datafile> <outfile>
The format of the parameter file <param> is:
dim <number>
obj <+|-> <+|-> ...
phi <number>
The output of bound to <outfile> is:
lower_bound <number> <number>... <number>
upper_bound <number> <number>... <number>
ideal <number> <number>... <number>
nadir <number> <number>... <number>
utopia_bound <number> <number>... <number>
nadir_bound <number> <number> ...<number>filter
A program that reads in a file consisting of a collection of approximation sets and outputs a collection of internally nondominated approximation sets. A separate parameter file gives the dimension of the data and whether each objective should be minimized or maximized.
./filter [<param>] <datafile> <outfile>
The format of the parameter file <param> is:
dim <number>
obj <+|-> <+|-> ...
method <0|1>normalize
A program that reads in a file, consisting of a collection of approximation sets, assumed to be from one optimizer. A separate parameter file gives the dimension of the data and whether each objective should be minimized or maximized. It also gives the upper and lower bound of each objective to normalize to. Finally, it specifies whether to unify all the objectives so that they are to be all minimized or all maximized, or not to unify them in this way. The output is the collection of points normalized so that they lie within the range 1,2 in each dimension and appropriately unified.
./normalize [<paramfile>] <boundfile> <datafile> <outfile>
The format of the parameter file <param> is:
dim <number>
obj <+|-> <+|-> ...
unify <min|max|no>
The bound file must contain two lines of the format:
upper_bound <number> <number>... <number>
lower_bound <number> <number>... <number>
Pipeline